New publication of Holger Dobbek and coworkers in PNAS
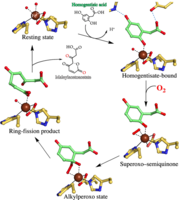
Life on a molecular scale depends on the constant formation and breakdown of metabolits. A variety of organisms ranging from soil bacteria to man employ the homogentisate pathway to degrade aromatic amino acids. Homogentisate is the last metabolite in the pathway with an aromatic ring, which needs to be broken by reacting with dioxygen, a reaction catalysed by the Fe2+-containing enzyme homogentisate dioxgenase. A lack of active homogentisate dioxygenase in humans causes alkaptonuria, a rare inherited genetic disorder.
Activated states of dioxygen make several different challenging reactions in the cell possible, but are also responsible for DNA mutations and cell aging. How homogentisate dioxgenase coordinates dioxygen activation with aromatic ring fission was unknown and has now been revealed in a crystallographic study conducted by Jae-Hun Jeoung, Martin Bommer, Tzong-Yuan Lin, and Holger Dobbek at the Humboldt-Universität zu Berlin.
In their recent paper in the early edition of PNAS (Proceedings of the National Academy of Sciences of the United States of America) they describe the crystal structures of five different steps along the reaction coordinate of homogentisate dioxygenase. Especially revealing are the structures showing how dioxygen coordinates Fe2+ to give a superoxo ligand facing a semiquinone radical formed from homogentisate, which combine in the next step to an alkylperoxo molecule in which the aromatic ring and dioxygen formed the first covalent bond, two central steps in the attack on the aromatic ring. The study was greatly aided by observing four different stages of the reaction in a single crystal.
Homoprotocatechuate dioxgenase, another Fe2+ containing enzyme featuring a fold unrelated to that of homogentisate dioxygenase was already earlier observed to favour aromatic ring cleavage through similar intermediates in an active site pocket of different architecture. However, the only structural similarities between both enzymes are the three protein ligands of Fe2+. Thus, the same minimal motif seems to be sufficient to direct reaction of dioxygen with a Fe2+-bound aromatic substrate – an observation encouraging biomimetic approaches to develop O2-activating catalysts.
See also:
Selected-Research-Highlights or
online at PNAS.