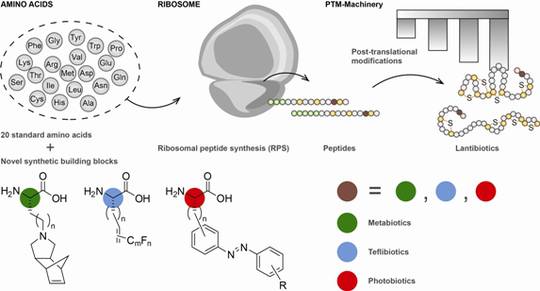
Biosynthetic machineries for the generation of peptide antibiotics
The central topic of this project is to use two highly complex multistep catalytic peptide and peptide biosynthesis assembly lines and to harness their biosynthetic capabilities to generate novel compounds. Apart from a deeper understanding of these biosynthesis machineries, new structural diversity will be generated as exemplified by pharmacologically active peptides, peptide derivatives and proteins.
In this approach, metabolic pathway engineering and combinatorial biosynthesis is intimately combined with innovative strategies in synthetic chemistry to generate specifically designed building blocks (D4). These modifications in the biosynthetic catalytic machineries will result in chemically diversified and unique antibiotic peptides and with built-in bioorthogonal chemical handles to modify the products post-biocatalytically.
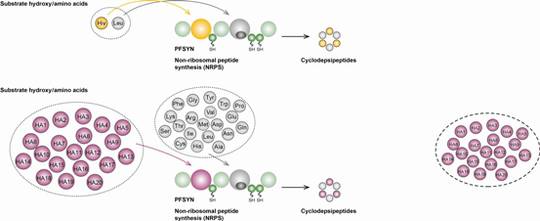
Bioactive peptides are synthesised from several microorganisms (e.g. Actinobacteria, Cyanobacteria, Bacilli, fungi) by two different types of biocatalytic assembly line, the ribosome (RPS) and the non-ribosomal peptide synthetases (NRPS).
Both biosynthetic assembly lines, RPS and NRPS, commonly rely upon an extensive spectrum of post-translational (RPS) and post-tailoring (NRPS) modifications including cyclisations, oxidations, halogenations, glycosylations and biaryl formations, expanding the structural diversity of the biosynthetic products.
Two models are chosen for these studies: lantibiotics (LANs, lanthionine containing antibiotics) with antibacterial properties from Gram-positive organisms such as Bacillus which are ribosomally synthesised, and CDPs, non-ribosomally synthesised from fungi with a variety of antifungal, insecticide, or antiviral bioactivities. Representatives of both structural classes are marketed drugs.
Research goals
A primary goal is to deepen the understanding of biosynthetic enzymes and their mechanisms by spectroscopic studies and determining the three-dimensional structures of specific enzymes of these machineries.
In parallel, manipulating and engineering of biosynthetic machineries with non-canonical amino acids will lead to new structural and functional diversity. The ultimate goal is to generate novel peptide antibiotics for potential biotechnological applications in the pharmaceutical and crop protection industries.
The transfer of optimised fermentation protocols to larger scale will be exlored. Selected lantibiotics produced in sufficiently large quantities (mg – g range) will subsequently be used for further bio-orthogonal chemical modifications.
A fundamental understanding of RPS and NRPS will be obtained by biochemical, structural, and spectroscopic investigations of the modular entities of NRPSs and components of the RPS. We will also employ IR, EPR, and fluorescence spectroscopic approaches, monitoring reporter groups that are introduced via site-specific incorporation of non-natural amino acids via orthogonal pairs.