B3 Cofactor insertion and functional investigations on complex molybdoenzymes
The molybdenum cofactor (Moco) is the essential component of a group of redox enzymes which are of major importance in human metabolism. Sulfite oxidase (SO) is essential for humans and catalyses the oxidation of sulfite to sulfate, the last step in the degradation of sulfur-containing amino acids. Xanthine dehydrogenase (XDH) and aldehyde oxidase (AO) are complex metallo-flavoproteins that contain Moco, two [2Fe2S] clusters and flavin adenine dinucleotide (FAD) as catalytically acting units, and catalyse the oxidative hydroxylation of purines, pyrimidines, pterines, and aldehyde substrates using NAD+ or molecular oxygen as electron acceptors [1]. The project centres on the mechanism of cofactor insertion and enzyme assembly and the spectroscopic characterisation of diverse molybdoenzymes.
While the biochemical function of XDH is well established, the biochemical and physiological role of AO is still obscure. AO is believed to play an important role in the metabolism of drugs and xenobiotica. To elucidate the role of this enzyme in the mammalian organism and to define the specific physiopathological function, AO isoforms (mouse, human, E. coli) were purified. The studies are paralleled by analysing similarities and differences of the active site structures and mechanisms of AO isozymes using various spectroscopic (IR, resonance Raman, EPR, XAS) and electrochemical techniques. The studies are based on the recent progress achieved for the purification of mammalian AOs in larger amounts, and extending the scope of the research programme to include interdisciplinary approaches and novel methods developed within the Cluster will help lead to an understanding of the complex enzyme systems of molybdo-flavoenzymes in detail.
Results/Achievements
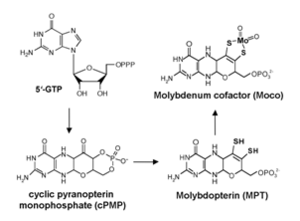
Our project was mainly focused on two parts: i) to characterise the molybdoenzyme aldehyde oxidase in detail with spectroscopic methods, electrochemistry and x-ray crystallography ii) to analyse the mechanism of electron transfer, assembly and cofactor insertion into different molybdoenzymes.
Part 1: Characterisation of the molybdoenzyme aldehyde oxidase
In this part we characterised different aldehyde oxidases from mouse, humans, aldehyde oxidoreductase from E. coli and sulfite oxidase from humans.
The most important highlight of this project was the identification of the molybdopterin cytosine dinucleotide cofactor (MCD) in the aldehyde oxidoreductase PaoABC from E. coli. The MCD cofactor was not expected to be present in E. coli. We investigated its biosynthesis and insertion into PaoABC [2].
Another highlight of the project was to obtain crystals of mAOH1 in order to solve the X-ray structure of mAOH1 in collaboration with the University of Lisbon (Prof. Maria Romao) [3].
Currently we are developing a biosensor for cinnamaldehyde (known to cause contact allergies) by using E. coli PaoABC. The electrochemic properties of PaoABC were so far investigated by direct protein voltammetry and mediated bioelectrocatalysis using different redox mediators.
In addition, human sulfate oxidase, a member of the sulfate oxidase family, was characterised by electrochemical methods. The enzyme was used for the development of a sulfite biosensor [4+5]. Further, sulfite oxidase was characterised by its redox properties and its catalytic activity using surface enhanced resonance Raman (SERR) spectroscopy and cyclic voltammetry (CV) [6]. A homology model for hSO was also constructed using as template the structure of chicken liver SO [7]. For the rational design, synthesis and optimization of nanoporous electrodes for enzyme and Moco immobilization for functional bioelectronic devices are currently used.
The mechanism of subtrate oxidation by aldehyde oxidases is currently analysed by theoretical calculations using quantum chemical methods (TD-DFT, QM/MM).
Part 2: Analysing the mechanism of electron transfer, assembly and cofactor insertion
We analysed the mechanism of electron transfer, assembly and cofactor insertion into different molybdoenzymes (Rhodabacter capsulatus XDH, E. coli YedY). In R. capsulatus XDH, amino acid substitutions at two cysteine residues coordinating FeSI of the two [2Fe-2S] clusters of the enzyme demonstrate that an incomplete assembly of FeSI impairs the formation of the XDH (ab)2 heterotetramer and, thus, insertion of Moco into the enzyme [8]. The FeS centers were characterised by EPR spectroscopy [9]. In addition, a specific protein involved in the insertion of Moco into RcXDH was described, named XdhC. The characterisation of XdhC will help to understand the mechanism of cofactor insertion into molybdoenzymes by enzyme-specific chaperones [10].
To analyse the mechanism of Moco maturation in more detail, and how the cofactor is inserted into target proteins, EXAFS studies were performed. The ligands at the Mo site of Moco were analysed in YedY [11]. These studies will help to understand how the molybdenum atom is coordinated in different molybdoenzymes. Model compounds for Moco were developed in an external collaboration with Trinity College Dublin (Prof. Carola Schulzke) [12].
Important publications
- Leimkühler, S., Wuebbens, M.M., and Rajagopalan, K.V. (2011) The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. CCR: in press.
- Neumann M, Mittelstädt G, Iobbi-Nivol C, Saggu M, Lendzian F, Hildebrandt P, Leimkühler S. (2009) A periplasmic aldehyde oxidoreductase represents the first molybdopterin cytosine dinucleotide cofactor containing molybdo-flavoenzyme from Escherichia coli. FEBS J. 276:2762-74.
- Mahro M, Coelho C, Trincao J, Rodrigues D, Terao M, Garattini E, Saggu M, Lendzian F, Hildebrandt P, Romao MJ, Leimkühler S. (2011) Characterisation and crystallization of mouse Aldehyde Oxidase 3 (mAOX3): from mouse liver to E. coli heterologous protein expression. Drug Metab Dispos. 2011 [Epub ahead of print].
- Spricigo, R., Richter, C., Leimkühler, S., Gorton, L. Scheller, F.W., Wollenberger, U. (2010) Sulfite biosensor based on osmium redox polymer wired sulfite oxidase. Colloids and Surfaces A: Physicochemical and Engineering Aspects 354: 314-319.
Spricigo, R., Dronov, R., Lisdat, F., Leimkühler, S. Scheller, F.W., and Wollenberger, U. (2009) Electrocatalytic sulfite biosensor with human sulfite oxidase coimmobilized with cytochrome c in a polyelectrolyte containing multilayer. Anal. Bioanal. Chem. 393: 225-33.
- Sezer M, Spricigo R, Utesch T, Millo D, Leimkühler S, Mroginski MA, Wollenberger U, Hildebrandt P, Weidinger I (2010) Redox properties and catalytic activity of surface-bound human sulfite oxidase studied by a combined surface enhanced resonance Raman spectroscopic and electrochemical approach. Phys Chem Chem Phys. 12:7894-903.
- Tillmann Utesch and Maria Andrea Mroginski (2010) Three-Dimensional Structural Model of Chicken Liver Sulfite Oxidase in its Activated Form. J. Phys. Chem Lett. 1, 2159–2164.
- Schumann, S., Saggu, M., Möller, N., Anker, S.D., Lendzian, F., Hildebrandt, P., and Leimkühler, S. (2008) The Mechanism of Assembly and Cofactor Insertion into Rhodobacter capsulatus Xanthine Dehydrogenase. J. Biol. Chem. 283:16602-11.
- Schumann S, Terao M, Garattini E, Saggu M, Lendzian F, Hildebrandt P, Leimkühler S. (2009) Site directed mutagenesis of amino acid residues at the active site of mouse aldehyde oxidase AOX1. PLoS One. 4:e5348.
- Neumann M, Leimkühler S. (2011) The role of system-specific molecular chaperones in the maturation of molybdoenzymes in bacteria. Biochem Res Int. 2011:850924.
- Havelius, K.G., Reschke, S., Horn, S., Döring, A., Niks, D., Hille, R., Schulzke, C., Leimkühler, S., Haumann, M. (2011) Structure of the Molybdenum Site in YedY, a Sulfite Oxidase Homologue from Escherichia coli. Inorg. Chem. in press.
- Prinson P. Samuel, Sebastian Horn, Alexander Döring, Kajsa G. V. Havelius, Stefan Reschke, Silke Leimkühler, Michael Haumann, Carola Schulzke (2011) A crystallographic and Mo K-edge XAS study of molybdenum-oxo bis-, mono-, and non-dithiolene complexes: First-sphere coordination geometry and non-innocence of ligands. Eur. J. Inorg. Chem. In press.
Project team and expertise
| Dr. Anna Fischer (TU Berlin) | Nanoporous electrodes for enzyme and Moco immobilization |
| Prof. Dr. Peter Hildebrandt (TU Berlin) | Vibrational spectroscopy, metalloenzymes, EPR spectroscopy |
| Prof. Dr. Silke Leimkühler (Uni Potsdam) | Molybdoenzymes, biochemistry |
| Prof. Dr. Maria-Andrea Mroginski (TU Berlin) | Vibrational spectroscopy |
| Prof. Dr. Thomas Risse (FU Berlin) | EPR Spectroscopy |
| Prof. Dr. Peter Saalfrank (Uni Potsdam) | Qantum chemical methods (TD-DFT, QM/MM) |
| Prof. Dr. Inez Weidinger (TU Berlin) | Raman spectroscopy |
| Prof. Dr. Ulla Wollenberger (Uni Potsdam) | Electrochemistry, Biosensors |
| PD Dr. Michael Haumann (FU Berlin) former member | EXAFS |
| Dr. Axel Warsinke (Uni Potsdam) former associated member | Biosensors |


