C2 Biocatalysis and process techniques
State of the art/Introduction
Peptides and polyketides from bacteria and fungi display various biological activities, e.g. antibacterial, antiviral, cytostatic etc., which are of great interest for their application as drugs in pharma or for crop protection. These compounds are numerous and quite diverse in structure, as their mechanisms of action are, which makes them useful for the treatment of infections, immune diseases, cardiovascular dysfunction and cancer.
Peptide antibiotics produced by bacterial or fungal microorganisms are either ribosomally synthesized (RPS), e.g. nisin (food preservative), ziconotide (analgesic), or, by non-ribosomal peptide synthetases (NRPS), e.g. vancomycin (antibiotic), bleomycin (cancer drug). A crucial point is the understanding of the biosynthesis of natural products on a genetic and enzymatic level as a prerequisite for the optimisation of production or the structural alteration of the natural products.
While ribosomal synthesis can be manipulated by site-directed mutagenesis or module shuffling, the NRPSs display a modular assembly of catalytic domains which, need to be exchanged.
In a synthetic biology approach, we perform the design and construction of new enzyme variants by combinatorial genetics and combinatorial biosynthesis. From the exchange of these domains, the generation of new structural diversity in the metabolite is expected. Although much of the structural and functional basis of the mechanisms of these multifunctional enzymes has been elucidated, the engineering of available NRPSs and the design of new robust enzymes from existing modules is still in its infancy. Finally, concepts of the design of new proteins are investigated in a basic research approach. This includes the generation of artificial proteins and their modulation of enzymatic and spectroscopic properties.
Research goals
- mechanistic characterization of posttranslational modifications in ribosomal peptide synthesis (RPS) and NRPS
- transfer of natural product biosynthesis from chemoenzymatics (in vitro) to combinatorial biosynthesis and mutasynthesis (in vivo) in heterologous hosts, e.g. E. coli
- establishing a library of modified NRPS modules and domains as a toolbox for the construction of new recombinant enzymes and the design of artificial enzymes and enzyme cascades for the generation of new structural diversity
- bioprofiling of new natural products for their bioactivity
Results/Achievements
In biological catalysis, apart from a mechanistic understanding, a central objective is the in vitro generation of novel diversity of natural products. In this research initiative the focus is laid on peptides produced by bacteria and fungi, displaying various interesting bioactivities. We have initially focused on the non-ribosomal peptide synthesis (NRPS) assembly line. The challenge was the manipulation, re-design and re-direction of the biosynthetic capabilities towards new peptide products. Model systems were the NRPS-based enzymes Enniatin-synthetase (ESYN) and PF synthetase (PFSYN) from fungi which both constitute assembly lines of progenitors of marketed drugs, i.e. the antibacterial Locabiosol® and the anthelmintic Emodepside®. The latter has the potential to be used as an agent against river blindness affecting several millions of people in less developed countries. In this context, considerable success could be achieved in the in vitro generation of new cyclodepsipeptides [1] and re-cently a breakthrough has been achieved for the in vivo expression and antibiotic production by such a NRPS in E. coli (unpublished data). This paves the way for combinatorial biosyn-thesis with NRPS (Figure 1).
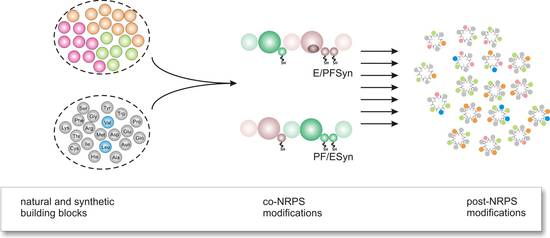 |
The successful heterologous expression of ribosomally synthesized peptide (RPS) antibiotics (Figure 2) [2] prompted us to include a second line of research, i.e. the RPS of lantibiotics [2, 5, 6]. These lantibiotics, originally synthesized by Gram-positive Bacillus and Actinomyces strains, display various bioactivities, e.g. acting against other Gram-positive bacteria. The remarkable finding of a heterologous expression in the Gram-negative host E. coli constitutes a paradigm change and spurs new hopes in the facile generation of new peptide variants with enhanced or altered bioactivities [5, 6]. In future, we will, therefore, employ advanced molecular biology techniques combined with innovative chemical catalytic protocols.
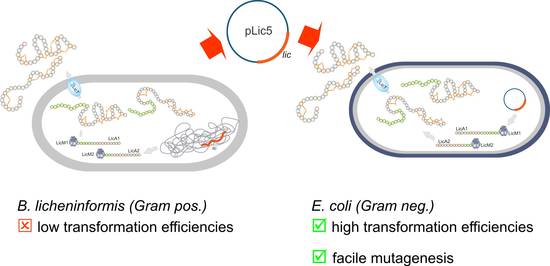 |
The manipulation of ribosomal peptide synthesis will lead to the incorporation of non-proteinogenic amino acids into proteins. Corresponding amino acids will be provided by sci-entists from research area E4. Hence, new chemical functionalities will be delivered by (i) global substitution of a single amino acid type [E40] or (ii) position-specific insertion of novel synthetic amino acids [4]. The basic prerequisite for site-directed insertions will be the use of orthogonal aminoacyl-tRNA synthetase:tRNA (aaRS:tRNA) pairs in the translation process. Five of these pairs are already available whereas additional pairs have to be developed during the funding period [4]. These additional functionalisations are considered as a basis for further chemical reactions (post-biosynthetic chemistry) provided by research field E4.
Most important publications
- In vitro synthesis of new cyclodepsipeptides of the PF 1022-type – probing the α-D-hydroxy acid tolerance of PF 1022-synthetase; Müller, J., Feifel, S.C., Schmiederer, T., Zocher, R., Süssmuth, R.D. ChemBioChem 2009 10, 323-328.
- Heterologous expression, biosynthesis and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli; Caetano, T., Krawczyk, J., Mösker, E., Süssmuth, R.D., Mendo, S., Chemistry & Biology 2011, 18, 1-11.
- Expanding and engineering the genetic code in a single expression experiment; Hoesl, M.G., Budisa, N. ChemBioChem 2011, in press.
- In Vivo Double and Triple Labeling of Proteins Using Synthetic Amino Acids; Lepthien, S., Merkel, L., Budisa, N. Angew. Chem. Int. Ed. 2010, 49, 5446-5450. Angew. Chem. 2010, 122, 5576-5581.
- In vitro Biosynthesis of the Prepeptide of Type-II Lantibiotic Labyrinthopeptin A2 Including Formation of a C-C-Bond as a Post-Translational Modification; (Hot paper). Müller, W., Schmiederer, T., Ensle, P., Süssmuth, R.D., Angew. Chemie Int. Ed. 2010, 49, 2436-2440.
- Labyrinthopeptins: A New Class of Carbacyclic Lantibiotics (VIP paper). Meindl, K. Schmiederer, T., Schneider, K., Reicke, A., Butz, D., Keller, S., Nicholson, G., Gühring, H., Vertesy, L., Wink, J., Hoffmann, H., Brönstrup, M., Sheldrick, G.M., Süssmuth, R.D., Angew. Chem. Int. Ed. 2010, 49, 1151-1154.
C2 Project team and expertise
| Prof. Dr. Nediljko Budisa (TU Berlin) | Synthetic biology and molecular biotechnology of proteins |
| PD Dr. Ullrich Keller (TU Berlin) Associated member | Biosynthetic enzymes of natural products |
| Prof. Dr. Rudibert King (TU Berlin) | Fermentation optimisation, modelling |
| Prof. Dr. Peter Neubauer (TU Berlin) | Biotechnology of enzymes in industrial processes |
| Prof. Dr. Roderich Süssmuth (TU Berlin) | Combinatorial biosynthesis, synthetic biology and biological chemistry of natural products |
Former team members | |
| Prof. Dr. Marion Ansorge-Schumacher (TU Berlin) | Hydrogenase biocatalysis |
| Prof. Dr. Siegfried Blechert (TU Berlin) | Substrate and natural product synthesis |
| Prof. Dr. Bärbel Friedrich (HU Berlin) | Hydrogenase characterisation |
| Prof. Dr. Matthias Kraume (HU Berlin) | Continuous screening system for biocatalysis |


